Genome Annotation
Genome Designs offers manual functional re-annotation of sequenced genomes to gain the highest possible number of encoded functions. This process dramatically improves the number of recognized functions mostly from the pool of numerous 'Putative Unrecognized Proteins' or unequivocal generic assignments generated by automatic annotations. The improvement also includes unique information on multifunctional "promiscuous" proteins.
The re-annotation standardizes protein function names and assigns EC numbers to secure correct mapping of sequenced genomes onto the best-fitting set of metabolic pathway diagrams from the MPW, our proprietary metabolic pathway database. Such re-annotation is required for improving comparative analysis of closely related genomes.
Functional Reconstruction
The central part of the Genome Designs' technology is Metabolic and Functional Reconstruction Platform. Originally developed by Genome Designs' CSO Dr. Evgeni Selkov in the beginning of the nineties, it allows predicting metabolic and regulatory networks based on a complete list of assigned gene functions, and a list of functions binary connected for all known variants of metabolic, transport, and signal transduction pathways organized as a metabolic pathway database.
MPW - Metabolic Pathway Database
Genome Designs' reconstruction technology critically depends on the underlying database of metabolic pathways and reactions. The MPW database we will be using for this project once made a revolution in microbial genomics, used as a foundation for the partially sequenced genomes reconstruction technology. The MPW database started through human curation back in 1990s, and now lists the majority of known pathway variations through its rather unique definition of a pathway as a chain of reactions between metabolic crossroads.
In recent years, the MPW database has been extensively updated in the private industry, and now contains the encoding of metabolism for all major domains of life. Lately, particular attention has been given to encoding metabolism of higher eukarya, including plants and human.
Data Analysis and Genome Annotation Software
Our modular data analysis and genome annotation suite allows us to flexibly adapt to a wide range of projects. Below is a diagram for our methodology of determining subsets of genes specifically expressed in each human tissue.
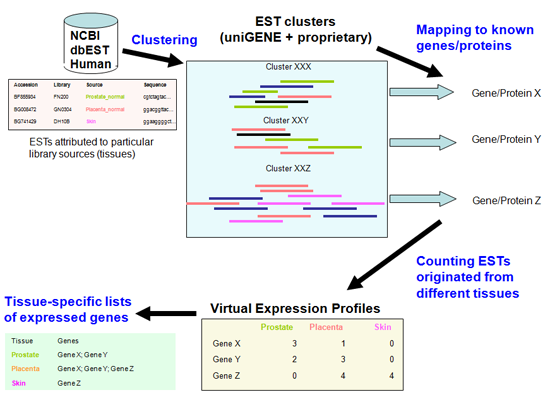
Creating tissue-specific expressed gene lists from a transcriptome analysis.
The Transcriptomics Data Analysis Pipeline takes collections of reads and performs quality and composition-based trimming, repeat detection and masking, chimera detection, clustering and mapping onto reference genomes, annotation of transcripts and construction of virtual expression profiles.
Other Software Tools
Genome Designs partners with SciDM Group, a bioinformatics software developer, to augment its existing genome annotation pipelines and to boost custom software development projects for clients. The following packages are being extensively used:
- SciDM DBMS - high performance zero-maintenance object-relational NoSQL database engine
- EMBEDB - embedded data access library serving as a back-end for SciDM System (open source)
- QSimScan - ultra-high speed DNA and protein sequence similarity search tool (open source)
Email us for more information.
Detailed information on each of these steps is available upon request.
Email us for more information.